STA410 Assignment 4
Michal Malyska
library(tidyverse)
library(mixtools)
knitr::opts_chunk$set(cache = TRUE)
buffalo <- scan("buffsnow.txt")Question 1
a)
It is very straightforward to see that:
\[ \theta_k = \theta_1 + (\theta_2 - \theta_1) + (\theta_3 - \theta_2) + \dots + (\theta_k - \theta_{k-1})\]
\[ \text{Since each of the brackets is just } \phi_{j} \]
\[ \theta_1 + \sum_{i=2}^{k}\phi_i \]
b)
We compute partial derivative for \(g(\theta)\) \[ \frac{\partial g(.)}{\partial {\theta_1}} = -2 \sum_{i = 1}^n \left( y_i - \theta_1 + \sum_{j=2}^i \phi_j \right) \] \[ \text{Setting to zero: } \frac{\partial g(.)}{\partial {\theta_1}} = 0 \] \[ \text{We obtain: } \theta_1 = \frac{1}{n} \sum_{i = 1}^k \left(y_i-\sum_{j=2}^i\phi_j \right) \]
c)
\[ \frac{\partial g(.)}{\partial {\phi_j}} \left( \sum_{i = 1}^n \left(y_i - \theta_1 - \sum_{j=2}^i \phi_j \right)^2 + \lambda \sum_{j = 2}^n | \phi_j | \right) = -2 \sum_{i = j}^n \left(y_i - \theta_1 - \sum_{k=2}^i \phi_k \right) +\lambda \frac{\partial | \phi_j | }{\partial {\phi_j}} \]
Where the \(\frac{\partial | \phi_j | }{\partial {\phi_j}}\) will be replaced by a subgradient of the absolute value.
\[ \lambda \frac{\partial | \phi_j | }{\partial {\phi_j}} = \begin{cases} \lambda & \phi_j > 0 \\ [- \lambda, \lambda] & \phi_j = 0 \\ - \lambda & \phi_j < 0 \end{cases} \]
Since this is zero only on the set of \([-\lambda, \lambda]\), we know that the overall function is going to be zero only this set as well. Applying that constraint we see:
\[ -2 \sum_{i = j}^n \left(y_i - \theta_1 - \sum_{k=2}^i \phi_k \right) \in [-\lambda, \lambda] \\ \iff \\ \left| \sum_{i = j}^n \left(y_i - \theta_1 - \sum_{k=2}^i \phi_k \right) \right| < \frac{\lambda}{2} \]
Now, if \(\phi_j \neq 0\) we can see that:
\[ \phi_j = \frac{1}{n-j+1} \left( \sum_{i = j}^n \left(y_i - \theta_1 - \sum_{k=2, k \neq j}^i \phi_k \right) - \frac{\lambda}{2} \right) \mbox{for} \sum_{i = j}^n \left(y_i - \theta_1 - \sum_{k=2, k \neq j}^i \phi_k \right) > \frac{\lambda}{2}\]
and the opposite case:
\[ \phi_j = \frac{1}{n-j+1} \left( \sum_{i = j}^n \left(y_i - \theta_1 - \sum_{k=2, k \neq j}^i \phi_k \right) + \frac{\lambda}{2} \right) \mbox{for} \sum_{i = j}^n \left(y_i - \theta_1 - \sum_{k=2, k \neq j}^i \phi_k \right) < \frac{\lambda}{2}\]
Question 2
a)
Let \(\theta\) represent the vector of parameters.
First we want to find the loglikelihood: \[ \log(L(\theta)) = \sum_{i=1}^n \sum_{k=1}^m u_{ik} \log f_k(x_i) + \sum_{i=1}^n \sum_{k=1}^m u_{ik} \log(\lambda_k) \]
It is only the right sum that depends on the lambda, and solving this problem under the constraint that \(\sum_{k=1}^{m}\lambda_k = 1\) It’s easy to see that: \[ \hat{\lambda_k} = \frac{1}{n}\sum_{i=1}^n u_{ik} \]
Similarily, plugging in the Normal density for \(f_k(.)\) and solving for \(\mu\) and \(\sigma\) we obtain the other two results:
\[ \frac{\partial l(\theta)}{\partial \mu_k} = \frac{1}{\sigma^2_k} \sum_{i=1}^{n}u_{ik}(x_i - \mu_k) \]
\[ \frac{\partial l(\theta)}{\partial \sigma^2_k} = - \frac{1}{2 \sigma_k^2} \sum_{i=1}^{n}u_{ik} + \frac{1}{2(\sigma^2_k)^2}\sum_{i=1}^{n}u_{ik}(x_i - \mu_k)^2 \]
Setting both to 0 and solving produces:
\[ \hat{\mu_k} = \frac{\sum_{i=1}^n u_{ik} x_i}{\sum_{i=1}^n u_{ik}} \]
\[ \hat{\sigma_k^2} = \frac{\sum_{i=1}^n u_{ik} (x_i - \hat{\mu}_k)^2}{\sum_{i=1}^n u_{ik}} \]
b) Implement the EM algorithm for the model above.
normalmixture <- function(x,k,mu,sigma,lambda,em.iter=50, eps = 10^-5, debug = FALSE) {
n <- length(x)
# Add a tiny perturbation to x
x <- x + rnorm(length(x), mean = 0, sd = eps)
x <- sort(x)
vars <- sigma^2
means <- mu
lam <- lambda/sum(lambda)
delta <- matrix(rep(0,n*k),ncol = k)
loglik <- NULL
converged = FALSE
iter = 0
while (!converged) {
loglik_new <- 0
for (i in 1:n) {
xi <- x[i]
for (j in 1:k) {
mj <- means[j]
varj <- vars[j]
denom <- 0
for (u in 1:k) {
mu <- means[u]
varu <- vars[u]
denom <- denom + lam[u]*dnorm(xi,mu,sqrt(varu))
}
delta[i,j] <- lam[j]*dnorm(xi,mj,sqrt(varj))/denom
}
loglik_new <- loglik_new + log(denom)
}
for (j in 1:k) {
deltaj <- as.vector(delta[,j])
lambda[j] <- mean(deltaj)
means[j] <- weighted.mean(x = x, w = deltaj)
vars[j] <- weighted.mean((x - means[j])^2, w = deltaj)
}
# Standardize lambdas
lambda <- lambda / sum(lambda)
lam <- lambda
# Iterate again
iter <- iter + 1
# Print iterations for debugging
if (debug) {cat("Iteration: ", iter, "\n", "Log Likelihood: ", loglik_new, "\n")}
# Update the first log likelihood
if (is.null(loglik)) {
if (debug) {print("loglik empty reached")}
loglik = loglik_new
}
# Check for convergence
else if (abs(tail(loglik, n = 1) - loglik_new) < eps) {
if (debug) {print("convergence reached")}
converged <- TRUE
}
# Check for num interations
if (iter == em.iter) {
print("Number of iterations reached")
converged <- TRUE}
# If likelihood is infinite then print the params
if (is.infinite(loglik_new)) {
cat("Infinite Likelihood", "\n")
r <- list(mu = means,
var = vars,
delta = delta,
lambda = lambda,
loglik = loglik,
iterations = iter)
return(r)
}
# Update the log likelihood
loglik <- c(loglik, loglik_new)
}
# return
r <- list(mu = means,
var = vars,
delta = delta,
lambda = lambda,
loglik = loglik,
iterations = iter)
return(r)
}c)
df <- tibble(x = buffalo)
df %>% ggplot(aes(x = x, ..density..)) +
geom_density(bw = 5, fill = "red", alpha = 0.3) +
labs(title = "Density plot")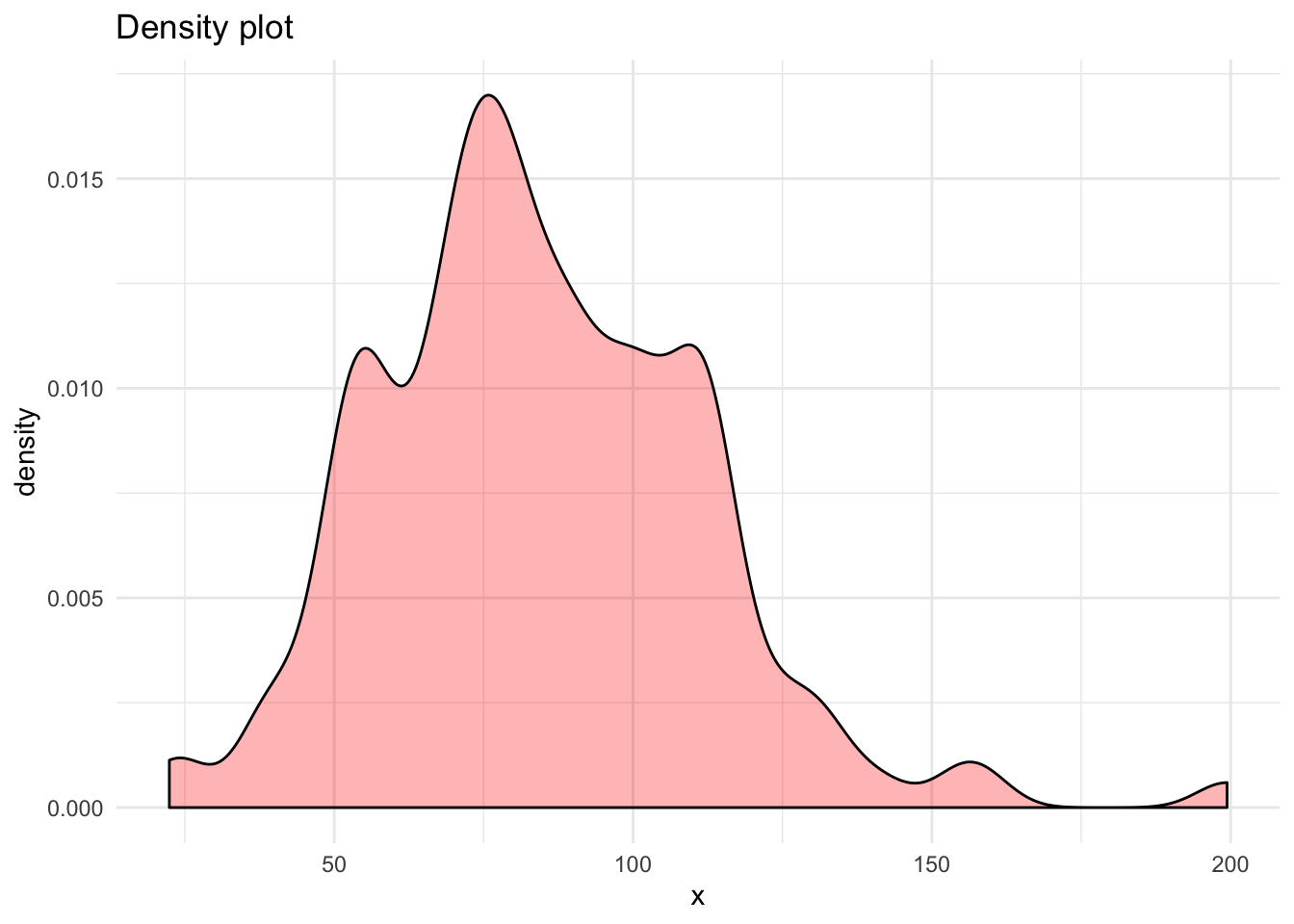
# From density plot:
breaks2 <- c(95)
breaks3 <- c(65, 105)
# Split into normals:
df <- df %>% mutate(bin2 = if_else(x < breaks2[1], 1, 2),
bin3 = if_else(x < breaks3[1], 1,
if_else(x < breaks3[2], 2, 3)))
# Calculate the means and variances of each component normals:
norms2 <- df %>% group_by(bin2) %>% summarize(mu = mean(x),
sd = sqrt(var(x)),
count = n())
norms3 <- df %>% group_by(bin3) %>% summarize(mu = mean(x),
sd = sqrt(var(x)),
count = n())
# run this on m = 2
mu_start <- as.vector(norms2$mu)
var_start <- as.vector(norms2$sd)
lambda_start <- as.vector(norms2$count) / length(df$x)
density2 <- normalmixture(df$x, 2, mu_start, var_start, lambda_start, em.iter = 1000, debug = FALSE)
# run this on m = 3
mu_start <- as.vector(norms3$mu)
var_start <- as.vector(norms3$sd)
lambda_start <- as.vector(norms3$count) / length(df$x)
density3 <- normalmixture(df$x, 3, mu_start, var_start, lambda_start, em.iter = 1000, debug = FALSE)
# Create comparison plot against standard density estimate
density1 <- density(df$x)
## new data:
df2 <- tibble( x = seq(from = min(df$x), to = max(df$x), length.out = 10000))
df2$density2 <- density2$lambda[1] * dnorm(df2$x, mean = density2$mu[1], sd = sqrt(density2$var[1])) +
density2$lambda[2] * dnorm(df2$x, mean = density2$mu[2], sd = sqrt(density2$var[2]))
df2$density3 <- density3$lambda[1] * dnorm(df2$x, mean = density3$mu[1], sd = sqrt(density3$var[1])) +
density3$lambda[2] * dnorm(df2$x, mean = density3$mu[2], sd = sqrt(density3$var[2])) +
density3$lambda[3] * dnorm(df2$x, mean = density3$mu[3], sd = sqrt(density3$var[3]))
# Make the plot:
ggplot(data = df2, aes(x = df2$x)) +
geom_point(aes(y = df2$density2), color = "red") +
geom_point(aes(y = df2$density3), color = "blue") +
geom_density(data = df, mapping = aes(x = x), fill = "green", alpha = 0.5) +
labs(main = "Density Estimates at m = 2 (red) and m = 3 (blue)")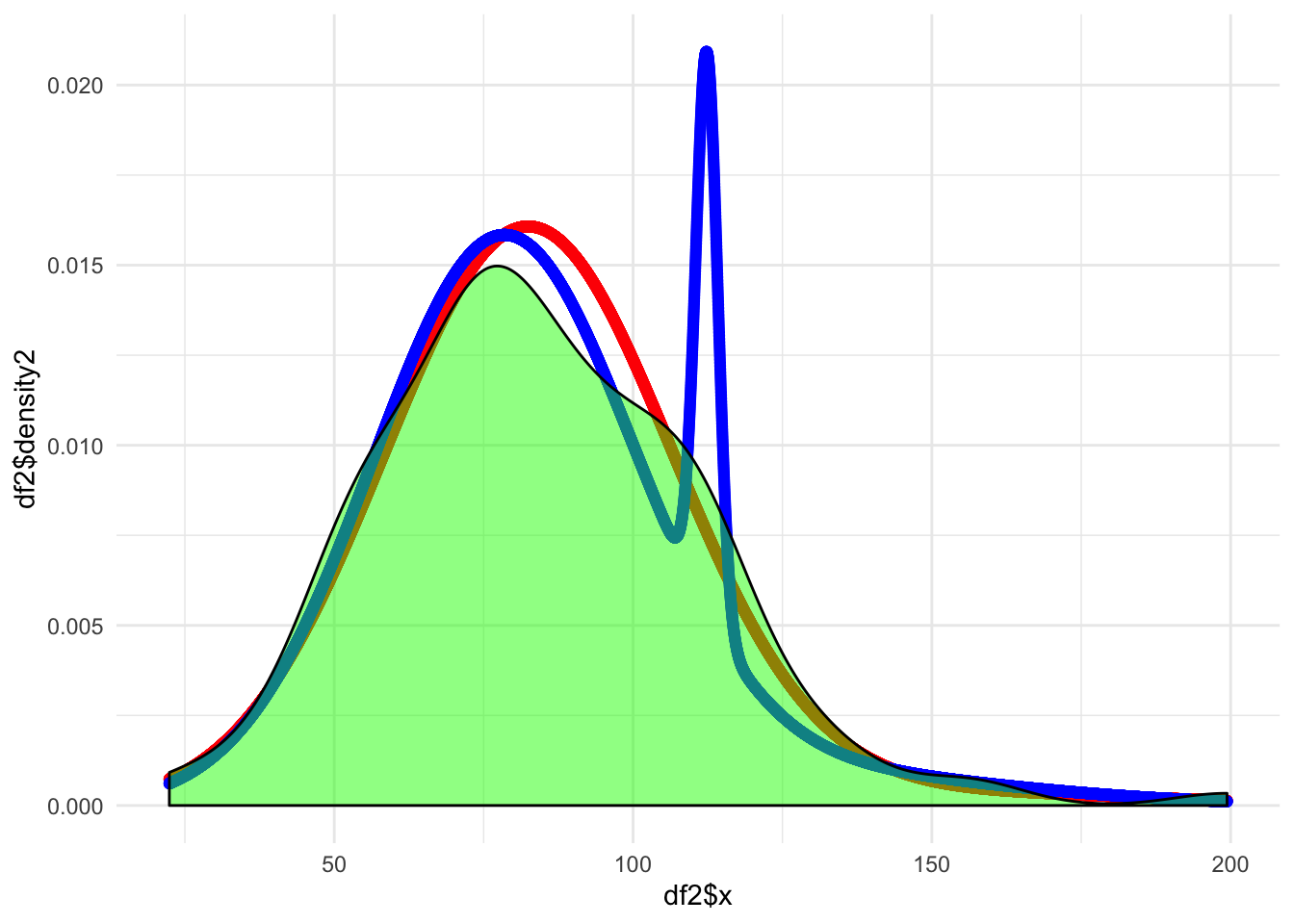
This tiny “hand” looks weird so I will do a sanity check with the mixtools package to see if my work seems reasonable.
mu_start <- as.vector(norms2$mu)
var_start <- as.vector(norms2$sd)
lambda_start <- as.vector(norms2$count) / length(df$x)
density4 <- mixtools::normalmixEM(df$x, k = 2,
mu = mu_start,
sigma = var_start,
lambda = lambda_start)## number of iterations= 431mu_start <- as.vector(norms3$mu)
var_start <- as.vector(norms3$sd)
lambda_start <- as.vector(norms3$count) / length(df$x)
density5 <- mixtools::normalmixEM(df$x, k = 3,
mu = mu_start,
sigma = var_start,
lambda = lambda_start)## number of iterations= 609print("Mixture of two parameter comparison (mine vs mixtools)")## [1] "Mixture of two parameter comparison (mine vs mixtools)"cat("Mean \n",
density2$mu, "\n",
density4$mu, "\n")## Mean
## 82.33512 150.314
## 82.41374 153.7458cat("Variance \n",
density2$var, "\n",
density4$sigma^2, "\n")## Variance
## 581.1774 1173.335
## 583.5366 1079.92cat("Lambda \n",
density2$lambda, "\n",
density4$lambda, "\n")## Lambda
## 0.9681888 0.03181124
## 0.970787 0.02921304print("Mixture of three parameter comparison (mine vs mixtools)")## [1] "Mixture of three parameter comparison (mine vs mixtools)"cat("Mean \n",
density3$mu, "\n",
density5$mu, "\n")## Mean
## 77.98207 112.3513 120.9678
## 78.01955 112.3522 121.6513cat("Variance \n",
density3$var, "\n",
density5$sigma^2, "\n")## Variance
## 471.8441 3.386749 1403.283
## 473.245 3.383962 1396.149cat("Lambda \n",
density3$lambda, "\n",
density5$lambda, "\n")## Lambda
## 0.8340415 0.07175839 0.09420006
## 0.836254 0.0716751 0.09207088Looks good to me.
d)
Comparison by AIC:
\[ AIC = 2k-2loglikelihood \]
for \(m = 2\) we have \(2\) means, \(2\) variances, and \(1\) mixing parameter (since the other one is just 1- the first one) for a total of \(k = 5\).
for \(m = 3\) we get \(k = 8\)
Then AIC for both models is:
\[ AIC_2 = 10 + 2 \cdot 628.5453 = 1267.091 \]
\[ AIC_3 = 16 + 2 \cdot 624.3508 = 1264.702 \]
Since the AIC has pretty much no difference we can say that both are very comparable. However given the shape of the plots I would prefer the mixture of 2 as it seems to be a much more smooth curve. The fit is also significantly faster.